The Wilson Research Laboratory at Georgia Institute of Technology - School of Chemical and Biomolecular Engineering, seeks to engineer novel, non-natural biological systems of bespoke function for high impact applications — while advancing our fundamental knowledge of protein and genetic structure-function relationships. The Wilson lab leverages a unique blend of iterative protein engineering and genetic engineering to design novel synthetic biological systems (i.e., biomolecular systems engineering). Our current focus is in the area of engineering cooperative systems of functional proteins and cognate genetic elements. To accomplish our goals, we employ workflows that involve rational protein design, synthetic biology, computational modeling, and laboratory evolution. These studies represent the most rigorous test of our understanding of structure-function and phylogenetic relationships; in addition to, promoting the development of novel biological tools that will benefit society.
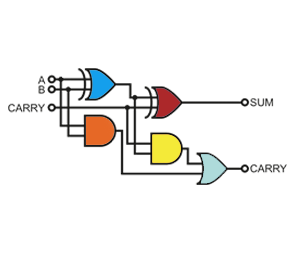
Project 1
Building a Biological Programming Structure
Description
The control of gene expression is an important tool for metabolic engineering, the design of synthetic gene networks, gene-function analysis, drug discovery and biopharmaceutical manufacturing. Several strategies have been developed to regulate gene expression and target protein production at different biosynthetic levels. The most widely used regulatory elements focus on blocking or activating mRNA synthesis by inducible coupling of transcriptional repressors or activators to constitutive or minimal promoters. Our long-term goal is to rationally design a new class of proteins that regulate gene expression, in which we can confer modulated DNA binding function that is responsive to specific exogenous cues and logical operations. This goal will require the complementary understanding and design of allosteric communication and functional molecular interactions (i.e., protein-ligand, protein-protein, and protein-DNA). To date we have engineered a set of novel transcription factors – i.e., repressors and antithetical anti-repressors. Likewise, we have engineered the corresponding genetic architectures to direct our transcription factors to non-synonymous promoters resulting in a marked increase in biological computing capacity. In addition to expanding our biological programming structure, we are currently applying this technology to a myriad of new applications across biotechnology.
Project 2
Engineering Intelligent Biological Systems
Description
Human stem cells have emerged as one of the most exciting resources in biotechnology, promising to revolutionize the treatment of heart disease, diabetes in addition to other diseases that require organ and tissue replacement. Stem cells have the unique ability to become specialized cells (e.g., muscle, neurological, immune) through a process called differentiation. Stem cells (or progenitor cells) differentiate into specific cell types with disparate functions via timing and order of transcription factor activation. In contrast, bacteria cells lack the ability to differentiate, thus cannot be induced to become specialized. However, through iterative protein and genetic engineering the edifice for the development of synthetic microbial “stem cells” can be realized. The engineering workflow to confer microbial differentiation will require the development of a synthetic memory structure to facilitate long term storage of information, and the design of complementary decision-making genetic programs that will enable the activation of bespoke genetic networks. The specific goal of this project is to create synthetic microbial stem cells. This objective will require the development of a genetic MEMORY structure that can rapidly rearrange genetic control infrastructures, resulting in genetic structural changes that persist in forward generations of the cell line - even after the initiation stimuli are removed. In addition, this goal will require the development of a dynamic synthetic transcriptional programming edifice that can generate two or more different combinational logical operations (i.e., programs) that are differentially responsive to INPUT signal combinations, facilitating two or more non-synonymous states of specialization. Successful completion of the proposed work will result in a significant paradigm shift in biomolecular systems engineering, and the development of schema and workflows for scalable genetic MEMORY structures and dynamic systems programming. The resulting biomolecular systems engineering edifice will revolutionize synthetic biology and metabolic engineering via the production of bespoke microbial progenitor cells that can evolve on cue, producing two or more states of disparate specialization.

Project 3
Developing Next-Generation Biological Security & Bio-Cryptography Tools
Description
The goal of this research is to engineer intelligent biological systems, with advanced security features. From an engineering perspective, an intelligent biological system has three fundamental tenets: (i) the ability to make decisions, (ii) a memory system that is coupled to decision making, and (iii) a means to communicate with other discrete living units. Engineered intelligent biological cells will enable the development of living therapeutics, advanced biomanufacturing capabilities, and contribute to a deeper understanding of complex functions in living systems. In addition, this collection of emerging technologies will enable next-generation biosecurity measures that will prevent the unwanted release of organisms that can cause harm to human life or property, and prevent the theft of valuable biotechnologies. In addition to the above, this research will contribute to workforce development in the areas of protein engineering, genetic engineering, and related technological fields. The successful completion of this research will result in several new chassis cells imbued with: (i) engineered programming structures for decision making, (ii) complementary memory structures to control states of specialization, and (iii) biosynthesis and receiver networks capable of programmable unidirectional and bidirectional communication. The resulting chassis cells will enable distributed computation, networked functions, expanded biological computing capabilities, and programmable differentiation. Moreover, the aforementioned technologies will enable the development of a broad range of novel bio-cryptography tools. Collectively this new biosecurity platform will enable scientists and engineers to progress beyond simple operational technology security measures, to a more robust informational technology intrinsic-security approach specifically designed for protecting biotic assets. To achieve these objectives will require the design and construction of novel gene controls forming a system of engineered allosteric transcription factors (i.e., transcriptional programming), a means to program correlated and permanent changes at the genetic level that can persist in forward generations of a given chassis cell, and the development of universal communication processes beyond standard quorum sensing.
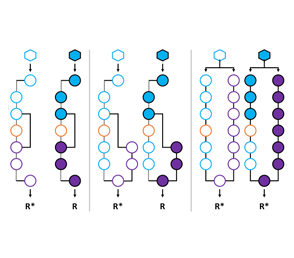
Project 4
Developing Design Rules for Allosteric Networks
Description
The means by which proteins communicate with their environment is critical to many life processes and will be required for the development of next-generation biotechnologies. Allosteric communication is the principal means by which proteins translate an environmental signal into a useful response (e.g., gene expression). Understanding the detailed mechanism of allostery is a vexing problem. Despite more than three decades of study this fundamental mode of biological communication remains unsolved at the molecular level. A thorough understanding of allosteric communication will facilitate the development of new protein design rules. The design of bespoke allosteric functions promises to revolutionize biotechnology, namely with regard to the development of more competent biological drugs, diagnostic tools, and industrial processes. These technological advances will be achieved by enabling precise and predictable biological communication in response to desired environmental cues. The goal of this study is to decipher the underlying molecular mechanism by which the allosteric signal traverses the scaffold across a number of engineered transcription variants with alternate allosteric control. This study will be accomplished by the construction and characterization of a synthetic phylogenetic tree of alternate allosteric networks. To complement inferred evolutionary relationships, experimental maps of communication will be constructed for two or more linages. Members of a given linage will be evaluated biophysically to decipher the underlying molecular mechanism. Phylogenetic and biophysical data will be leveraged to design alternate allosteric routes in certain engineered transcription scaffold in which we confer precise performance metrics (i.e., tuned dynamic range, ligand sensitivity and temporal responsiveness). This study will enable us to identify whether allostery in the LacI scaffold can be conferred via multiple networks of residues, or whether allostery requires a single conserved network of residues. Upon completion, this study will enable us to test our assertions with regard to the origin and molecular mechanics of alternate allosteric communication via the development of design rules for specific allosteric operations. In turn this algorithm will produce novel transcription factors for use in a broad range of biotechnological applications, specifically in the area of synthetic biology. In addition, this study will significantly broaden our fundamental understanding of allosteric communication.
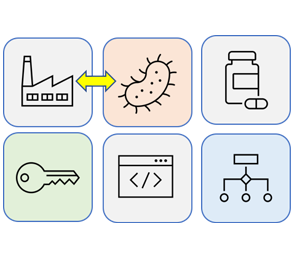
Project 5
Next-generation Biosynthesis & Biomanufacturing
Description
Modern biomanufacturing is a collection of unique technologies that utilize biological systems to construct commercially-relevant biomaterials and biomolecules (e.g., valuable products for use in medical, industrial, and defense applications). Biomanufactured products are found in natural sources like cultures of microbes, blood or plant and animal cells that have been artificially grown in specialized equipment. The cells used in the production may have occurred naturally or through advanced genetic engineering techniques. Biomanufacturing has enable the production of many important protein and small molecule products (e.g., Humulin, Taxol®). Moreover, metabolic engineering, as well as synthetic biology tools promise to revolutionize many aspects of biomanufacturing in terms of conferring improvements to (or development of novel) biological synthesis and intrinsic control (feedback systems) in vivo. In addition, bespoke electronic devices have emerged as promising technologies enabling the transform of biological inputs into non-biological outputs. We propose emerging biotic-abiotic manufacturing technologies, in which intrinsic (biotic) and electronic devices (abiotic) work cooperatively to achieve unprecedented (never seen before) control systems that will define biomanufacturing for the next three-decades.
Project 6
Growing Convergence Research: Biological, Electrical, & Chemical Engineering
Description
This Growing Convergence Research project aims at development of the emerging field of Biomolecular Systems Engineering. To accomplish this goal the team incorporates expertise from synthetic biology, protein engineering, process design, control theory and circuit design. The convergence of these disciplines will facilitate a radically new approach toward biological programming and the tools to accomplish these goals. The focus of the proposed work is to overcome reproducibility issues currently associated with the field of biological engineering by systemic organization and development of: i) engineering workflows, ii) process design tools, iii) curation and validation of standardized parts and iv), undergraduate and graduate pedagogy to advance the field of biological engineering. Successful completion of the proposed work will result in the creation of systematic workflows for the production and reporting of standardized biological unit operations and a new biological design software framework that will enable the production of scalable transcriptional programs, memory structures, and control systems for a variety of biological chassis. This award reflects NSF's statutory mission and has been deemed worthy of support through evaluation using the Foundation's intellectual merit and broader impacts review criteria.
Project 7
The Rational Design of Energy Transfer Protein Systems
+
Project 8
Generalized Concepts for the Engineering of Protein Structural and Functional Stability
+